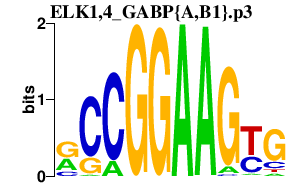
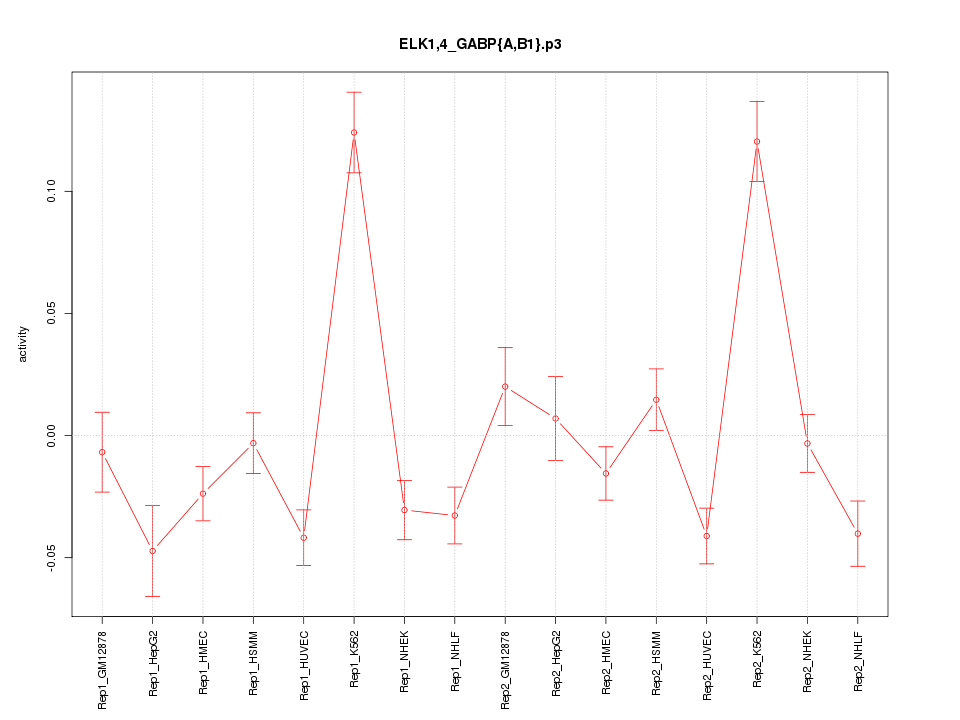
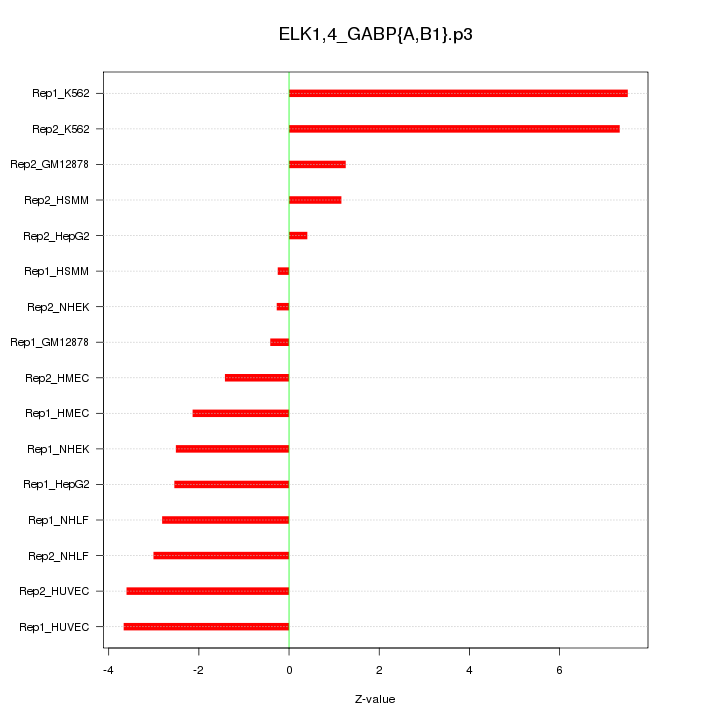
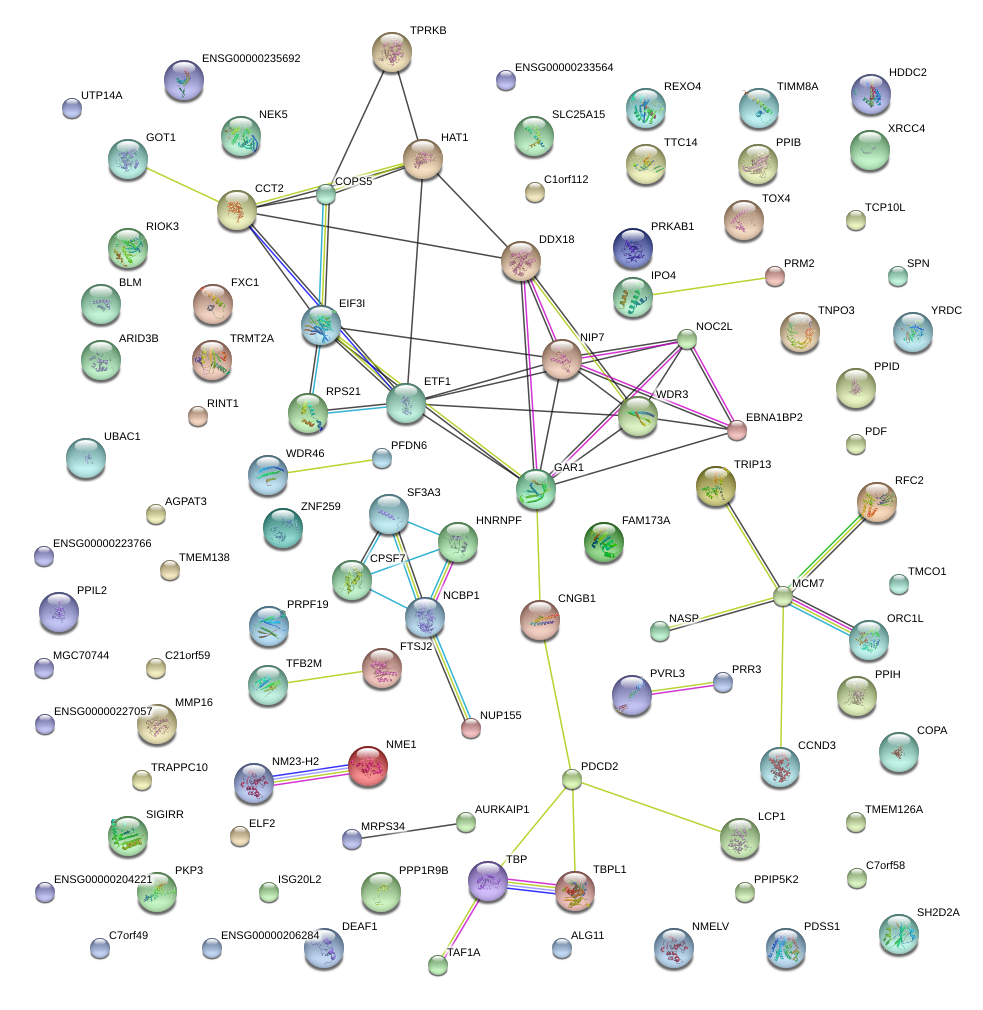
|
chr1_-_38273823
|
4.747
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr7_-_99698278
|
4.513
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr3_+_180319898
|
4.426
|
NM_001042601
NM_133462
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr4_+_110736636
|
4.174
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr21_-_33984865
|
4.083
|
NM_021254
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr1_-_38273683
|
3.806
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr12_+_120105656
|
3.741
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr1_-_38273846
|
3.655
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr7_-_128694823
|
3.642
|
|
TNPO3
|
transportin 3
|
|
chr7_-_2281797
|
3.415
|
|
FTSJ2
|
FtsJ homolog 2 (E. coli)
|
|
chr10_-_43892697
|
3.343
|
NM_001098207
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr1_-_38273852
|
3.241
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr6_+_170863420
|
3.228
|
NM_001172085
NM_003194
|
TBP
|
TATA box binding protein
|
|
chr9_-_138853146
|
3.158
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr7_-_128695118
|
3.035
|
|
TNPO3
|
transportin 3
|
|
chr9_-_138853007
|
3.026
|
|
UBAC1
|
UBA domain containing 1
|
|
chr1_-_222763081
|
2.992
|
NM_001201536
NM_005681
NM_139352
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa
|
|
chrX_-_100603956
|
2.940
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr4_-_159644439
|
2.932
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr13_-_46756326
|
2.908
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr21_-_33984657
|
2.861
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr1_-_38455636
|
2.848
|
NM_006802
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa
|
|
chr1_-_52870058
|
2.773
|
NM_001190818
NM_001190819
NM_004153
|
ORC1
|
origin recognition complex, subunit 1
|
|
chr5_+_892950
|
2.729
|
NM_001166260
NM_004237
|
TRIP13
|
thyroid hormone receptor interactor 13
|
|
chr13_-_46756295
|
2.718
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr10_-_43892675
|
2.707
|
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr1_-_246729546
|
2.681
|
NM_022366
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr1_-_894651
|
2.677
|
NM_015658
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae)
|
|
chr6_+_30524662
|
2.659
|
|
PRR3
|
proline rich 3
|
|
chr1_-_894620
|
2.645
|
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae)
|
|
chr7_-_134855383
|
2.566
|
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr7_-_2281822
|
2.494
|
NM_013393
|
FTSJ2
|
FtsJ homolog 2 (E. coli)
|
|
chr18_+_21033263
|
2.472
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr6_-_125623214
|
2.401
|
NM_016063
|
HDDC2
|
HD domain containing 2
|
|
chr1_-_1310817
|
2.376
|
NM_017900
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chr2_+_118572284
|
2.329
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr4_-_140005344
|
2.316
|
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr1_-_1310553
|
2.296
|
NM_001127230
NM_001127229
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chr7_-_134855237
|
2.294
|
NM_001243752
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr15_+_91260575
|
2.251
|
NM_000057
|
BLM
|
Bloom syndrome, RecQ helicase-like
|
|
chr16_+_771138
|
2.228
|
NM_023933
|
FAM173A
|
family with sequence similarity 173, member A
|
|
chr11_-_417324
|
2.202
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr1_+_169764174
|
2.191
|
|
C1orf112
|
chromosome 1 open reading frame 112
|
|
chr11_-_60673854
|
2.184
|
|
PRPF19
|
PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae)
|
|
chr20_+_60962120
|
2.177
|
NM_001024
|
RPS21
|
ribosomal protein S21
|
|
chr1_-_246729439
|
2.165
|
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr6_-_33257286
|
2.132
|
NM_001164267
NM_005452
|
WDR46
|
WD repeat domain 46
|
|
chr11_+_6502607
|
2.123
|
NM_012192
|
FXC1
|
fracture callus 1 homolog (rat)
|
|
chr7_-_128695181
|
2.104
|
NM_001191028
NM_012470
|
TNPO3
|
transportin 3
|
|
chr22_+_22020272
|
2.103
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr1_+_118472364
|
2.099
|
NM_006784
|
WDR3
|
WD repeat domain 3
|
|
chr16_-_1823104
|
2.091
|
NM_023936
|
MRPS34
|
mitochondrial ribosomal protein S34
|
|
chr16_+_29674564
|
2.082
|
|
SPN
|
sialophorin
|
|
chr9_+_100396079
|
2.078
|
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa
|
|
chr4_-_140005464
|
2.076
|
NM_006874
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr12_+_69979202
|
2.068
|
NM_006431
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr11_+_61129831
|
2.068
|
|
TMEM138
|
transmembrane protein 138
|
|
chr6_+_30525050
|
2.067
|
|
PRR3
|
proline rich 3
|
|
chr1_-_156698188
|
2.063
|
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2
|
|
chr7_-_134855530
|
2.049
|
NM_001243749
NM_001243751
NM_001243754
NM_024033
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr7_-_73668693
|
2.028
|
|
RFC2
|
replication factor C (activator 1) 2, 40kDa
|
|
chr10_+_26986557
|
2.028
|
NM_014317
|
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1
|
|
chr1_+_43124047
|
2.027
|
NM_006347
|
PPIH
|
peptidylprolyl isomerase H (cyclophilin H)
|
|
chr11_-_116658738
|
2.013
|
NM_003904
|
ZNF259
|
zinc finger protein 259
|
|
chr9_-_136282963
|
2.006
|
|
REXO4
|
REX4, RNA exonuclease 4 homolog (S. cerevisiae)
|
|
chr6_-_125623071
|
1.993
|
|
HDDC2
|
HD domain containing 2
|
|
chr2_+_118572254
|
1.968
|
NM_006773
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr22_-_20104428
|
1.937
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr17_+_49230929
|
1.923
|
|
NME1
|
non-metastatic cells 1, protein (NM23A) expressed in
|
|
chr12_+_69979268
|
1.922
|
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr1_+_46049659
|
1.916
|
NM_001195193
NM_002482
NM_152298
|
NASP
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr5_+_82373316
|
1.912
|
NM_003401
NM_022406
NM_022550
|
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4
|
|
chr11_-_116658701
|
1.905
|
|
ZNF259
|
zinc finger protein 259
|
|
chr2_+_172778907
|
1.882
|
NM_003642
|
HAT1
|
histone acetyltransferase 1
|
|
chr8_-_67974335
|
1.869
|
|
COPS5
|
COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis)
|
|
chr5_-_137878915
|
1.868
|
NM_004730
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr6_-_170893668
|
1.865
|
NM_001199462
NM_001199463
NM_001199464
NM_001199461
NM_002598
NM_144781
|
PDCD2
|
programmed cell death 2
|
|
chr15_+_74833517
|
1.859
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr1_-_160312959
|
1.848
|
|
|
|
|
chr6_+_33257408
|
1.848
|
NM_001185181
|
PFDN6
|
prefoldin subunit 6
|
|
chr2_+_172778973
|
1.842
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr7_+_105172531
|
1.831
|
NM_021930
|
RINT1
|
RAD50 interactor 1
|
|
chr1_-_160313056
|
1.831
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr6_-_33256946
|
1.830
|
|
WDR46
|
WD repeat domain 46
|
|
chr6_+_134274247
|
1.830
|
NM_004865
|
TBPL1
|
TBP-like 1
|
|
chr21_-_33984455
|
1.822
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr7_+_120629444
|
1.822
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr14_-_24658010
|
1.819
|
NM_024658
|
IPO4
|
importin 4
|
|
chr1_-_165737987
|
1.812
|
|
TMCO1
|
transmembrane and coiled-coil domains 1
|
|
chr11_-_61197272
|
1.802
|
NM_024811
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chr1_-_156786500
|
1.792
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr21_+_45432202
|
1.789
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr16_+_69373414
|
1.784
|
NM_001199434
NM_016101
|
NIP7
|
nuclear import 7 homolog (S. cerevisiae)
|
|
chr21_+_45285026
|
1.779
|
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr1_+_32688010
|
1.769
|
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chrX_+_129040076
|
1.766
|
NM_001166221
NM_006649
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast)
|
|
chr16_-_69373473
|
1.757
|
NM_032382
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr11_+_394238
|
1.753
|
|
PKP3
|
plakophilin 3
|
|
chr2_-_73964427
|
1.744
|
NM_016058
|
TPRKB
|
TP53RK binding protein
|
|
chr7_-_73668732
|
1.742
|
NM_002914
NM_181471
|
RFC2
|
replication factor C (activator 1) 2, 40kDa
|
|
chr2_+_172778961
|
1.734
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr1_-_43637788
|
1.733
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr5_+_102455957
|
1.724
|
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr6_+_33257350
|
1.724
|
NM_014260
|
PFDN6
|
prefoldin subunit 6
|
|
chr14_-_24657986
|
1.723
|
|
IPO4
|
importin 4
|
|
chr6_-_42016383
|
1.722
|
|
CCND3
|
cyclin D3
|
|
chr5_-_37371198
|
1.721
|
|
NUP155
|
nucleoporin 155kDa
|
|
chr7_-_91875296
|
1.714
|
NM_194455
NM_194456
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr22_+_19419341
|
1.712
|
|
MRPL40
|
mitochondrial ribosomal protein L40
|
|
chr2_+_172779000
|
1.708
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr6_-_33290678
|
1.700
|
NM_001141969
NM_001141970
NM_001350
|
DAXX
|
death-domain associated protein
|
|
chr8_-_131028646
|
1.698
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_+_32688053
|
1.697
|
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chr5_-_37371078
|
1.693
|
|
NUP155
|
nucleoporin 155kDa
|
|
chr17_-_40169400
|
1.684
|
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7
|
|
chr16_+_20817766
|
1.681
|
NM_001144924
NM_001199053
NM_030941
|
LOC81691
|
exonuclease NEF-sp
|
|
chr7_-_32534869
|
1.680
|
NM_001130710
NM_001139499
|
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_-_172413207
|
1.677
|
|
PIGC
|
phosphatidylinositol glycan anchor biosynthesis, class C
|
|
chr4_-_76439479
|
1.669
|
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr9_-_136283157
|
1.665
|
NM_020385
|
REXO4
|
REX4, RNA exonuclease 4 homolog (S. cerevisiae)
|
|
chr10_-_51623214
|
1.663
|
NM_006327
|
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast)
|
|
chr5_+_157158322
|
1.661
|
NM_017872
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae)
|
|
chr7_-_99006215
|
1.661
|
|
PDAP1
|
PDGFA associated protein 1
|
|
chr6_+_33257467
|
1.661
|
|
PFDN6
|
prefoldin subunit 6
|
|
chr7_-_91875108
|
1.659
|
NM_004912
NM_001013406
NM_194454
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr12_+_132195595
|
1.649
|
|
SFSWAP
|
splicing factor, suppressor of white-apricot homolog (Drosophila)
|
|
chr17_-_62493085
|
1.649
|
NM_007215
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit
|
|
chr7_-_45151272
|
1.645
|
|
TBRG4
|
transforming growth factor beta regulator 4
|
|
chr7_-_99006170
|
1.637
|
|
PDAP1
|
PDGFA associated protein 1
|
|
chr12_+_100661203
|
1.634
|
NM_017988
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr11_-_61197455
|
1.632
|
NM_001136040
NM_001142565
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chr19_-_56632644
|
1.631
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr17_-_40169130
|
1.629
|
NM_001144766
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7
|
|
chr16_-_20817771
|
1.624
|
NM_001142725
NM_080663
|
ERI2
|
ERI1 exoribonuclease family member 2
|
|
chr1_-_236767787
|
1.613
|
NM_018072
|
HEATR1
|
HEAT repeat containing 1
|
|
chr10_-_43892160
|
1.602
|
NM_001098208
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr17_+_49230896
|
1.602
|
NM_000269
NM_198175
NM_001018136
|
NME1
NME1-NME2
|
non-metastatic cells 1, protein (NM23A) expressed in
NME1-NME2 readthrough
|
|
chr19_-_50083813
|
1.595
|
|
NOSIP
|
nitric oxide synthase interacting protein
|
|
chr13_-_46756291
|
1.594
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr22_-_22901421
|
1.594
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr19_-_50083791
|
1.594
|
NM_015953
|
NOSIP
|
nitric oxide synthase interacting protein
|
|
chr18_+_21033230
|
1.584
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr19_+_36486042
|
1.576
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr1_+_55181494
|
1.568
|
NM_004623
|
TTC4
|
tetratricopeptide repeat domain 4
|
|
chr8_-_37707300
|
1.567
|
|
BRF2
|
BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like
|
|
chr11_-_47869901
|
1.566
|
NM_015231
|
NUP160
|
nucleoporin 160kDa
|
|
chr2_+_85839213
|
1.562
|
|
USP39
|
ubiquitin specific peptidase 39
|
|
chr20_-_44993000
|
1.562
|
NM_015945
NM_173073
NM_173179
|
SLC35C2
|
solute carrier family 35, member C2
|
|
chr16_+_20818010
|
1.561
|
|
LOC81691
|
exonuclease NEF-sp
|
|
chr17_-_17184589
|
1.558
|
NM_003653
|
COPS3
|
COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis)
|
|
chr4_+_41937105
|
1.558
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr1_+_32687970
|
1.557
|
NM_003757
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chr19_-_55791446
|
1.553
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr9_+_131218678
|
1.550
|
NM_153433
NM_153440
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr16_+_67261010
|
1.549
|
NM_014187
|
TMEM208
|
transmembrane protein 208
|
|
chr10_-_22292362
|
1.547
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr12_+_69080722
|
1.538
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr17_-_40169447
|
1.535
|
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7
|
|
chr11_-_60674039
|
1.532
|
NM_014502
|
PRPF19
|
PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae)
|
|
chr8_-_67974548
|
1.531
|
NM_006837
|
COPS5
|
COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis)
|
|
chr19_-_55791490
|
1.525
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr22_-_18257221
|
1.524
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr7_+_107384573
|
1.519
|
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr1_-_150552077
|
1.519
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr12_+_100660846
|
1.513
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr1_+_32688013
|
1.510
|
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chrX_-_153775427
|
1.510
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr10_+_12238179
|
1.509
|
|
CDC123
|
cell division cycle 123 homolog (S. cerevisiae)
|
|
chr1_-_47779651
|
1.501
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr19_+_10397642
|
1.495
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr2_-_27579277
|
1.494
|
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa
|
|
chr9_+_131218426
|
1.486
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr15_-_58358120
|
1.486
|
NM_001206897
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr10_+_105156382
|
1.485
|
NM_014976
|
PDCD11
|
programmed cell death 11
|
|
chr6_+_30585485
|
1.482
|
NM_014046
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr12_+_94071150
|
1.481
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr1_-_172413171
|
1.481
|
|
PIGC
|
phosphatidylinositol glycan anchor biosynthesis, class C
|
|
chr9_+_131218402
|
1.480
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr19_-_55791727
|
1.479
|
NM_001130106
NM_012267
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr2_+_177134122
|
1.476
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chrX_+_151885383
|
1.473
|
NM_153488
|
MAGEA2B
MAGEA2
|
melanoma antigen family A, 2B
melanoma antigen family A, 2
|
|
chr21_-_15918635
|
1.467
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr11_-_47869864
|
1.459
|
|
NUP160
|
nucleoporin 160kDa
|
|
chr5_-_37371182
|
1.459
|
NM_153485
|
NUP155
|
nucleoporin 155kDa
|
|
chr8_-_67974146
|
1.452
|
|
COPS5
|
COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis)
|
|
chr22_+_39898248
|
1.450
|
NM_019008
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr7_+_106505876
|
1.445
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr9_-_132597561
|
1.443
|
NM_016520
|
C9orf78
|
chromosome 9 open reading frame 78
|
|
chrX_-_151920098
|
1.443
|
NM_153488
|
MAGEA2B
MAGEA2
|
melanoma antigen family A, 2B
melanoma antigen family A, 2
|
|
chr11_-_64512925
|
1.437
|
NM_153819
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr19_+_36139221
|
1.432
|
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous)
|
|
chr17_+_80009775
|
1.425
|
|
GPS1
|
G protein pathway suppressor 1
|
|
chr1_+_161123755
|
1.421
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr17_+_49230938
|
1.417
|
|
NME1
NME1-NME2
|
non-metastatic cells 1, protein (NM23A) expressed in
NME1-NME2 readthrough
|
|
chr19_+_39421562
|
1.416
|
NM_021107
NM_033363
|
MRPS12
|
mitochondrial ribosomal protein S12
|
|
chr6_-_30585016
|
1.414
|
NM_002714
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr7_-_72722786
|
1.414
|
NM_001168347
NM_001168348
NM_018044
NM_148956
|
NSUN5
|
NOP2/Sun domain family, member 5
|
|
chrX_-_131351917
|
1.413
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr7_+_50348255
|
1.413
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr9_+_131218435
|
1.410
|
|
ODF2
|
outer dense fiber of sperm tails 2
|